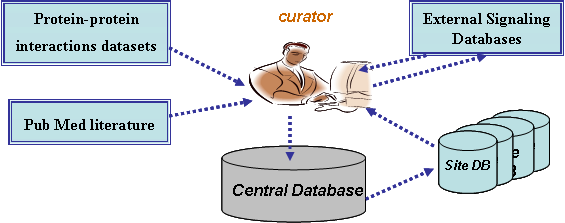
SPIKE stores a central DB containing carefully ascertained information uploaded by SPIKE users and supervised by the curators, data from external signaling DBs (e.g., KEGG, Reactome, IntAct), and wide-scale human protein-protein interaction datasets.
Each research lab that installs SPIKE (a.k.a. site) uses its own, local, database. During SPIKE's installation each site's database is populated with data from the central DB. Each site's database is periodically updated by the central DB. At any time, a site may choose to share parts of its data with the rest of the SPIKE community by uploading its private database to the central database.

The different types of biological entities are represented in SPIKE maps by nodes of different colors: violet nodes correspond to protein-coding genes, turquoise nodes to non protein-coding genes (e.g., micro-RNAs, rRNAs, tRNAs), green nodes to protein complexes, and yellow nodes to protein families. Small molecules are displayed in orange (not included in this map). Blue directed edges represent regulations: arrows correspond to activation, T shape edges to inhibition, and open circles denote regulations whose effect is still not clear (e.g., ATM was reported to phosphorylate MCM2, but the effect of this modification is not known yet). Blue undirected edges represent protein-protein interactions (not included in this map). Green edges represent containment relations between nodes (e.g., a complex and its components). Red and green dots within a node indicate that not all regulation and containment relations stored in the DB for the node are displayed in the map. This map represents the ATM-regulated network which is set off by the cell in response to double-strand breaks in the DNA.
Other features of the visualization software include:
- Superposition of high-throughput genomic and proteomic data.
- Linkage of information displayed in the maps to external DBs (e.g., Entrez Genes, PubMed).
- Display of paths between two genes.
- Display of all the direct connections within sets of genes.
- Finding signaling maps enriched with a set of genes.
- Filtering of the data displayed according to different parameters.