|
|
||||||||||||||
|
|
|
||||||||||||
|
†† Robust Inference in Dynamic
Bayesian Networks: †† Application to
Gene Expression Time-Series Data†††††††††
†††† Omer Berkman ††† Time series gene expression experiments
are very popular for studying a wide range of biological systems. We are
interested in the problem of inferring a genetic regulatory network from a
collection of temporal gene expression observations, in particular, with
small number of observations. Few approaches were suggested for this hard
problem and one of the most promising is the Dynamic Bayesian Network (DBN).
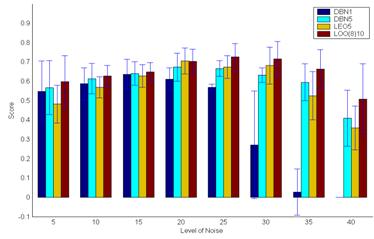
Several studies have investigated the robustness of the DBN estimation, based
on short time series of gene expression data, with the conclusion that robust
estimation of a genetic regulatory network, under mild simplifying
assumptions and without exploiting prior biologic knowledge, can be achieved
provided that the number of temporal observations is in the hundreds.
Unfortunately, this is unrealistic with current technologies, and raises the
need of better learning methods. †††† This project introduces a weak learnerís
methodology for this inference problem, studies few methods to produce Weak
Dynamic Bayesian Networks (WDBNs), and demonstrates their advantages
regarding the DBN model on simulated and real gene expression data. Our
results show that the novel algorithms produce more robust inference, with
respect to the number of observations and the level of noise within the
data.†
. |
|
|||||||||||||
|
|
||||||||||||||