|
 |
| Molecular Docking Algorithm
Based on Shape Complementarity Principles |
PatchDock is an algorithm for molecular docking. The
input is two molecules of any type: proteins, DNA, peptides, drugs.
The output is a list of potential complexes sorted by shape
complementarity criteria.
Short Overview:
PatchDock algorithm is inspired by object recognition and image segmentation
techniques used in Computer Vision. Docking can be compared to assembling a
jigsaw puzzle. When solving the puzzle we try to match two pieces by picking
one piece and searching for the complementary one. We concentrate on the
patterns that are unique for the puzzle element and look for the matching
patterns in the rest of the pieces. PatchDock employs a similar
technique. Given two molecules, their surfaces are divided into patches
according to the surface shape. These patches correspond to patterns that
visually distinguish between puzzle pieces. Once the patches are identified,
they can be superimposed using shape matching algorithms. The algorithm has
three major stages:
-
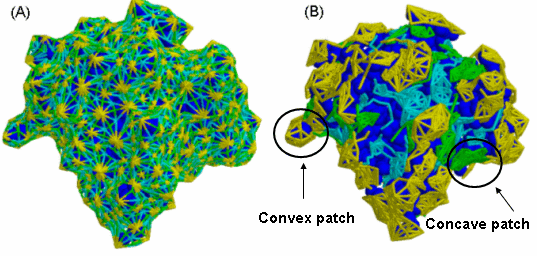
Molecular Shape Representation - in this step we compute the molecular surface
of the molecule. Next, we apply a segmentation algorithm for detection of
geometric patches (concave, convex and flat surface pieces). The patches are
filtered, so that only patches with 'hot spot' residues are retained.
-
Surface Patch Matching - we apply a hybrid of the Geometric Hashing and
Pose-Clustering matching techniques to match the patches detected in the
previous step. Concave patches are matched with convex and flat patches with
any type of patches.
-
Filtering and Scoring - the candidate complexes from the previous step are
examined. We discard all complexes with unacceptable penetrations of the atoms
of the receptor to the atoms of the ligand. Finally, the remaining candidates
are ranked according to a geometric shape complementarity score.
References:
- Duhovny D, Nussinov R, Wolfson HJ.
Efficient Unbound Docking of Rigid Molecules.
In Gusfield et al., Ed. Proceedings of the 2'nd Workshop
on Algorithms in Bioinformatics(WABI) Rome, Italy,
Lecture Notes in Computer Science 2452, pp. 185-200, Springer Verlag, 2002
[ PDF file ]
- Schneidman-Duhovny D, Inbar Y, Polak V, Shatsky M, Halperin I, Benyamini H, Barzilai A, Dror O, Haspel N, Nussinov R, Wolfson HJ.
Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking.
Proteins. 2003 Jul 1; 52(1): 107-12.
[ Abstract ]
[ PDF file ]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ.
PatchDock and SymmDock: servers for rigid and symmetric docking.
Nucl. Acids. Res. 33: W363-367, 2005.[ Full text ]
Beta 1.3 Version
Contact: duhovka@gmail.com