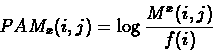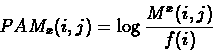Next: Distance between DNA Sequences
Up: Distance Matrix Methods
Previous: Pairwise Distances
Distance between Proteins - PAM matrices
We have already defined the PAM matrices, when we discussed heuristics for
sequence alignment (in lecture #3). The PAMn matrix is designed to compare two
amino-acid sequences which are n PAM units apart. Its calculation involves
raising M, the mutation probabilities matrix for one PAM unit, to the power of
n. For a continuous distance function, we need to define PAM matrices for non-
integer units, as well.
Let
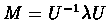 be the diagonalization of M, where
be the diagonalization of M, where  is a
diagonal matrix, made up of M's eigen-values, and U is an orthonormal matrix,
which consists of the corresponding eigen-vectors. Given a real x, the PAMx
distance matrix is simply:
is a
diagonal matrix, made up of M's eigen-values, and U is an orthonormal matrix,
which consists of the corresponding eigen-vectors. Given a real x, the PAMx
distance matrix is simply:
where f(i) is the frequency of the i-th amino-acid, and
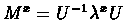 .
.
Itshack Pe`er
1999-02-18