


Next: Multi Experiment Analysis
Up: Analyzing Gene Expression Data
Previous: Introduction
Temporal Gene Expression Patterns
In a previous study [11], the authors established some
relationships between temporal gene expression patterns of 112 rat CNS
(Central Nervous System) genes and the development process of the rat's CNS.
Three major gene families were considered: Neuro-Glial Markers family (NGMs),
Neurotransmitter Receptors family (NTRs) and Peptide Signaling family (PepS).
All other genes measured in this study were lumped by the authors into a
fourth family: Diverse (Div). All families were further subdivided by the
authors, based on apriori biological knowledge. Gene expression patterns for
the 112 genes of interest were measured (using RT/PCR: [16]) in
cervical spinal cord tissue, at nine different developmental time points. This
yielded a
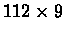 matrix of gene expression data. To capture the
temporal nature of this data, the authors transformed each (normalized)
9-dimensional expression vector into a 17-dimensional vector, including also
the 8 difference values between expression levels in successive time points.
This transformation emphasizes the similarity between genes with closely
parallel, but offset, expression patterns. Euclidean distances between the
augmented vectors were computed, yielding a
matrix of gene expression data. To capture the
temporal nature of this data, the authors transformed each (normalized)
9-dimensional expression vector into a 17-dimensional vector, including also
the 8 difference values between expression levels in successive time points.
This transformation emphasizes the similarity between genes with closely
parallel, but offset, expression patterns. Euclidean distances between the
augmented vectors were computed, yielding a
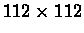 distance matrix.
Next, a phylogenetic tree was constructed for this distance matrix (using the
FITCH program [4]). Finally, cluster boundaries were determined by
visual inspection of the resulting tree. Some correlation between the
resulting clusters and the apriori family information was observed.
The CAST algorithm was tried on the same data in the following way: The raw
expression data was preprocessed in a similar manner: first the normalized
expression levels were augmented with the derivative values. Then, a
similarity matrix was computed based on the L1 distance between the
augmented 17-dimensional vectors. The CAST algorithm was applied to the
similarity matrix. Clusters were directly inferred
(figure 12.8).
distance matrix.
Next, a phylogenetic tree was constructed for this distance matrix (using the
FITCH program [4]). Finally, cluster boundaries were determined by
visual inspection of the resulting tree. Some correlation between the
resulting clusters and the apriori family information was observed.
The CAST algorithm was tried on the same data in the following way: The raw
expression data was preprocessed in a similar manner: first the normalized
expression levels were augmented with the derivative values. Then, a
similarity matrix was computed based on the L1 distance between the
augmented 17-dimensional vectors. The CAST algorithm was applied to the
similarity matrix. Clusters were directly inferred
(figure 12.8).
Figure 12.8:
The unprocessed data is compared
to the output of the clustering algorithm. Top: The similarity matrix of the
unprocessed data, compared against the new permutation according to the found
clusters. Bottom: The raw gene expression matrix is ordered according to the
permutation produced by the clustering algorithm and compared to the original
order.
|
|



Next: Multi Experiment Analysis
Up: Analyzing Gene Expression Data
Previous: Introduction
Itshack Pe`er
1999-03-16

