Next: Bibliography
Up: Analyzing Gene Expression Data
Previous: Temporal Gene Expression Patterns
Multi Experiment Analysis
Clustering gene expression patterns is useful even if the numbering of the
experiments has no physical meaning (as opposed to temporal patterns). Using
the CAST algorithm, data [1] for 1246 C. elegance genes, from 146
experiments, was analyzed. The data was in the form log(red/green)
(representing the log-ratio of the two sample intensity values at the
corresponding array feature), per experiment. Contrary to the first trial,
where the similarity measure needed to reflect the temporal nature of the
data, the order of experiments here, in the total set, has little or no
importance. Therefore, we use a unique similarity measure here.
Figure 12.9 summarizes the results. For temporal data it
makes sense to use other similarity measures when the corresponding sub
matrices are clustered. Clustering the columns (rather than the rows) of the
expression matrix is also possible and contains biologically meaningful
information.
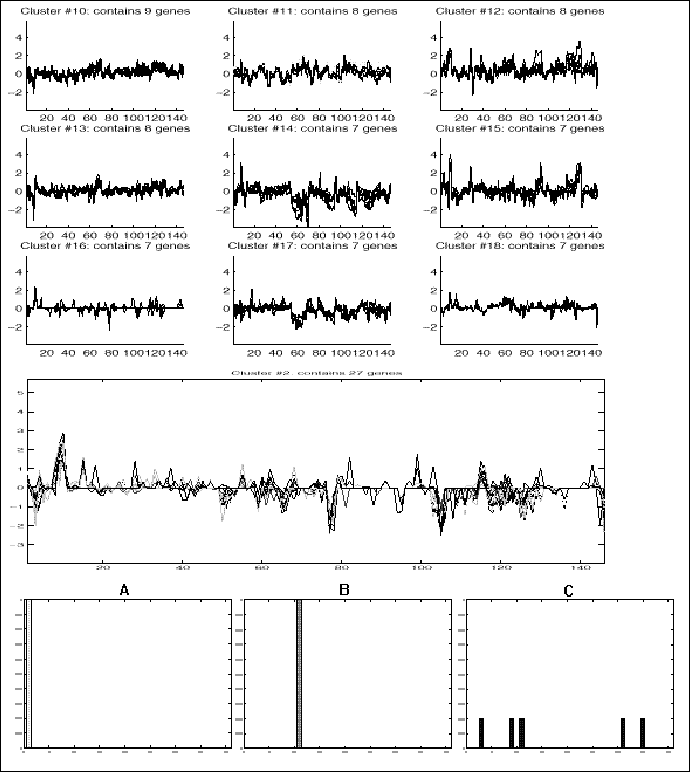
Figure 12.9:
Top: Examples of the clusters
found in analyzing the data for 1246 C. elegance genes. Below, cluster No. 2
has been enlarged. With a more precise version of this image, it is possible
to identify regions that significantly contribute to correlations within a
cluster, and then analyze the corresponding sub matrix. At the bottom, three
examples are given, presenting clustering results for different genes in the
same pre-defined family. Genes coding sperm proteins (8 genes, presented in
A) are all clearly grouped together. The same is true for dehydrogenase genes
(3 genes, presented in B). ATP related genes don't specifically correlate with
any other pattern. This is expected since ATP is involved in many cell
processes and is not correlated with specific conditions.
|
|



Next: Bibliography
Up: Analyzing Gene Expression Data
Previous: Temporal Gene Expression Patterns
Itshack Pe`er
1999-03-16