SPIKE user’s manual
3. Analysis utilities
This section explores how SPIKE
enhances
the analysis of functional genomics or
proteomics datasets.
Superposition of gene expression data
SPIKE enables the superposition of microarray data on top of the signaling maps. SPIKE
supports both absolute (e.g., Affymetrix chips) and relative (e.g., cDNA microarrays) gene
expression data. For further information regarding the expression data file format, see
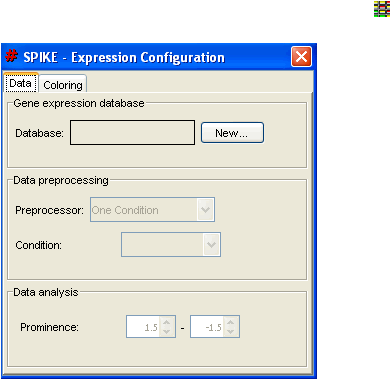
To start the analysis, select the "Gene Expression" button on the toolbar (
). The
expression configuration tool dialog opens.
To open the gene expression data file, select "New…" on the tool dialog. Next, select the
expression data file and its type (absolute or relative expression values; see Appendix A for
further details).
SPIKE shows for each displayed gene the fold change in expression between two selected
conditions, called 'test' and 'base'. Initially, all nodes will be colored either gray - indicating
that the dataset contains no relevant data concerning them (i.e., their corresponding gene is
not present on the microarray used in the experiment), or yellow - indicating no significant
fold change. This is because the test condition and the base condition are initially the same.
Change the "Test Condition" box value to another value. Now more colors should appear
on the map. Use the tooltip to learn the fold change values of some of the nodes.
SPIKE provides two alternative coloring schemes, 'Basic' and 'Graded'. The coloring of the
map can also be changed by setting the fold threshold values or the colors themselves.

