SPIKE user’s manual
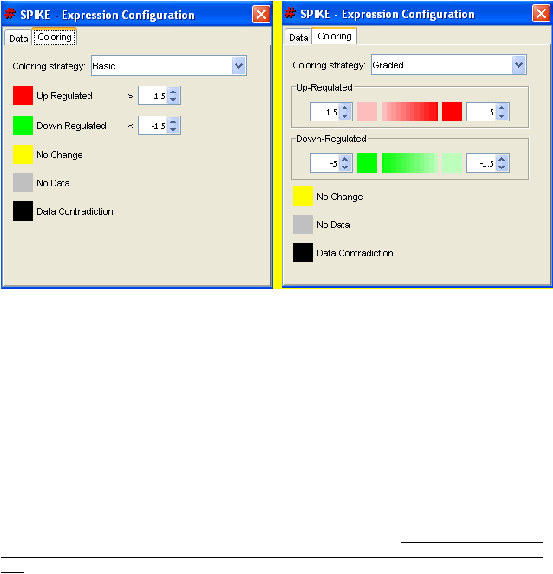
For example, with the "Basic" coloring strategy, default values are: genes whose fold
change was bigger than 1.5 will be colored red; genes whose fold change was smaller than
-1.5 (that is, decrease in expression by more than 1.5) will be colored green; those with
values in between will be colored yellow; and those with no available data are colored gray.
The "Data contradiction" coloring will be explained below.
The "Graded" coloring strategy enables users to match a range of fold change values to a
range of colors, thereby allowing them to differentiate between large and small fold changes
according to a scale of their choice. E.g., in the default settings, up-regulated values will be
colored from light-red (1.5) to dark-red, getting darker as the fold change increases, till 5.0
(values above 5.0 will be colored as 5.0).
The "Data" pane of the gene expression configuration tool dialog is composed of three
main panels:
The first panel deals with the gene expression data sources (files). Note that you may work
with multiple expression data files in a single session, switching back and forth between
files.
The second panel deals with some preprocessing of the data. In case the file contains
absolute values of expression levels (e.g., files generated by experiments that use
Affymetrix chips) only coloring of nodes according to a comparison between two conditions
(using the "Two Conditions" preprocessor) is possible. When working with a relative-type
file (i.e., files that contain relative expression levels obtained by comparing expression in
two conditions, e.g., files generated by experiments that use cDNA microarrays), coloring
according to the values of a single condition (using the "One Condition" preprocessor) is
also possible. The Threshold parameter is available for absolute-type files. Its default value
is 40. This means that all values in the file that are less than 40 are considered to be 40.
The threshold controls the sensitivity when comparing values one of which is very small.
The third panel deals with analysis of the data. In case the chip contains multiple probes for
certain genes, the program determines whether or not the data are contradictory. Data of a
gene are considered contradictory if there are probes of the same gene with values (fold
changes)
on both "sides" of the "Prominence" range (that is, for the same gene, some
probes indicate that the gene was up-regulated while some other indicate that it was down-
regulated). With the default settings, this would mean values both above 1.5 and below -1.5
(Note that these thresholds are independent of the thresholds indicating up-regulation or
down-regulation in the "Coloring" pane). For each gene whose values are not contradictory,
a single average value is calculated from all its probes and the gene's node is colored
accordingly. On the other hand, contradictory nodes are, by default, colored in black (see
the "Coloring" pane, and the tooltip of the expression color box would supply all the
contradictory values in that case.

