SPIKE user’s manual
Coloring Gene Clusters
SPIKE can superimpose any genes' partition information ("Clustering") on the map (see
Appendix A for further details on the clustering file format). The mechanism is similar to the
To activate the clustering view, select the "Clustering" button in the toolbar (
). This opens
the clustering tool dialog. Use the dialog to select the clustering file and type.
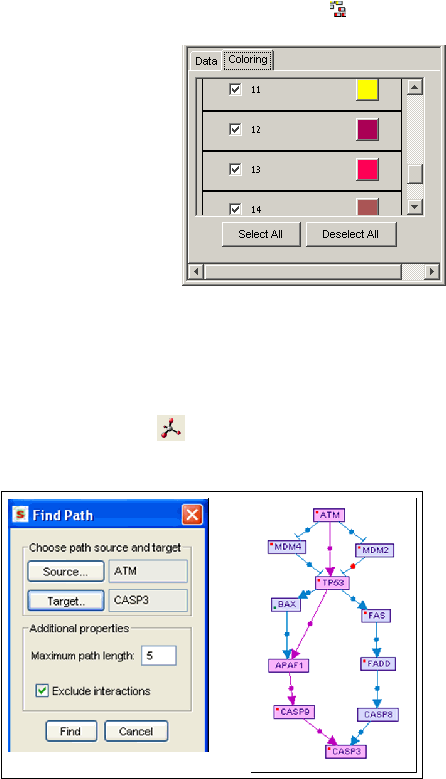
Move to the "Coloring" pane in the clustering
configuration dialog. You can select which
clusters to color, either with the checkbox on the
left of each cluster, or by selecting or de-
selecting all of them with the buttons on the
bottom. You can also change the colors that are
automatically applied to each cluster, by
pressing on the color button and changing its
settings.
Path Finding
SPIKE implements a path finding utility that finds and displays all directed paths, up to a
pre-defined maximal length, that connect source and target nodes selected by the user.
Selecting the Find Path button in the toolbar (
)
opens the corresponding tool dialog in
which the user determines the source and target nodes, as well as the maximal length for
the directed path connecting them. By default, un-directed edges (that is, protein-protein
interactions) are excluded from the analysis. All paths that meet the length constraint are
displayed with the
shortest one
highlighted.

